| Content: | 100 assays (50 of each) per kit |
| Shipping Temperature: | Ambient |
| Storage Temperature: |
Short term stability: 2-8oC, Long term stability: See individual component labels |
| Stability: | > 1 year under recommended storage conditions |
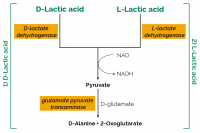
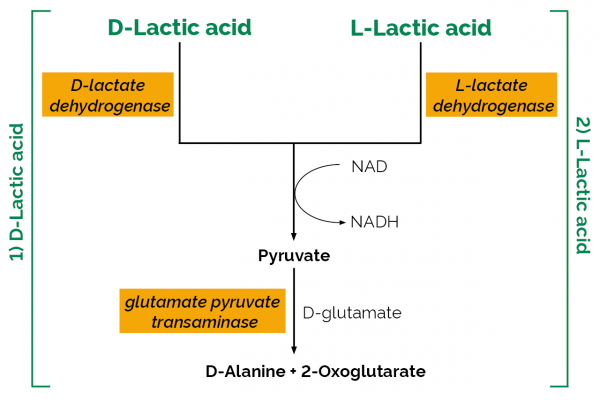
| Analyte: | D-Lactic Acid, L-Lactic Acid |
| Assay Format: | Spectrophotometer |
| Detection Method: | Absorbance |
| Wavelength (nm): | 340 |
| Signal Response: | Increase |
| Linear Range: | 0.5 to 30 µg of D- or L-lactic acid per assay |
| Limit of Detection: | 0.21 mg/L |
| Reaction Time (min): |
~ 10 min (L-lactic acid), ~ 5 min (D-lactic acid) |
| Application examples: | Wine, soft drinks, milk, dairy products, foods containing milk (e.g. dietetic foods, bakery products, baby food, chocolate, sweets and ice-cream), vinegar, fruit and vegetables, processed fruit and vegetables, meat products, food additives, paper (and cardboard), cosmetics, pharmaceuticals and other materials (e.g. biological cultures, samples, etc.). |
| Method recognition: | Methods based on this principle have been accepted by DIN, GOST, IDF, EEC, EN, ISO, OIV, IFU, AIJN and MEBAK |
The D-/L-Lactic Acid (D-/L-Lactate) (Rapid) test kit is used for the rapid and specific concurrent measurement and analysis of L-lactic acid (L-lactate) and D-lactic acid (D-lactate) in beverages, meat, dairy and food products.
Note for Content: The number of manual tests per kit can be doubled if all volumes are halved. This can be readily accommodated using the MegaQuantTM Wave Spectrophotometer (D-MQWAVE).
Explore more organic acid assay kit products.

- Extended cofactors stability. Dissolved cofactors stable for > 1 year at 4oC.
- Rapid total analysis time (concurrent / flexible D and L-lactic acid reaction format)
- D-lactate dehydrogenase reaction very rapid with most samples (~ 5 min)
- Very competitive price (cost per test)
- All reagents stable for > 2 years after preparation
- Mega-Calc™ software tool is available from our website for hassle-free raw data processing
- Standard included
Megazyme “advanced” wine test kits general characteristics and validation.
Charnock, S. J., McCleary, B. V., Daverede, C. & Gallant, P. (2006). Reveue des Oenologues, 120, 1-5.
Many of the enzymatic test kits are official methods of prestigious organisations such as the Association of Official Analytical Chemicals (AOAC) and the American Association of Cereal Chemists (AACC) in response to the interest from oenologists. Megazyme decided to use its long history of enzymatic bio-analysis to make a significant contribution to the wine industry, by the development of a range of advanced enzymatic test kits. This task has now been successfully completed through the strategic and comprehensive process of identifying limitations of existing enzymatic bio-analysis test kits where they occurred, and then using advanced techniques, such as molecular biology (photo 1), to rapidly overcome them. Novel test kits have also been developed for analytes of emerging interest to the oenologist, such as yeast available nitrogen (YAN; see pages 2-3 of issue 117 article), or where previously enzymes were simply either not available, or were too expensive to employ, such as for D-mannitol analysis.
Hide AbstractGrape and wine analysis: Oenologists to exploit advanced test kits.
Charnock, S. C. & McCleary, B. V. (2005). Revue des Enology, 117, 1-5.
It is without doubt that testing plays a pivotal role throughout the whole of the vinification process. To produce the best possible quality wine and to minimise process problems such as “stuck” fermentation or troublesome infections, it is now recognised that if possible testing should begin prior to harvesting of the grapes and continue through to bottling. Traditional methods of wine analysis are often expensive, time consuming, require either elaborate equipment or specialist expertise and frequently lack accuracy. However, enzymatic bio-analysis enables the accurate measurement of the vast majority of analytes of interest to the wine maker, using just one piece of apparatus, the spectrophotometer (see previous issue No. 116 for a detailed technical review). Grape juice and wine are amenable to enzymatic testing as being liquids they are homogenous, easy to manipulate, and can generally be analysed without any sample preparation.
Hide AbstractUnveiling the impact of traditional sourdough propagation methods on the microbiological, biochemical, sensory, and technological properties of sourdough and bread: a comprehensive first study.
Pontonio, E., Perri, G., Calasso, M., Celano, G., Verni, M. & Rizzello, C. G. (2025). Applied Food Research, 5(1), 101037.
In the last thirty years, the factors driving the establishment and composition of the sourdough biota have been deeply studied. Nevertheless, to date, no study has ever evaluated the biochemical and microbial dynamics of sourdoughs propagated using the different traditional methods integrated into procedural back-slopping practices worldwide. A mature type I sourdough was propagated for 10 days according to four managing conditions (Milanese, In Water, Free and Piedmontese) entailing incubations in a jute sack, submerged in water, in a jar or a combination of them. Sourdoughs obtained under the different conditions (and corresponding breads) were extensively characterized. When processing parameters modified the sourdough environment, the microbial community changed. In the first days of propagation Fructilactobacillus sanfranciscensis was the main dominant species regardless of the type of propagation, remaining present in all sourdoughs, especially those maintained in a jar. Differences among the propagation methods emerged from the biochemical analysis. Sourdoughs propagated in water exhibited higher titratable acidity, mainly due to the acetic acid produced, and were characterized by a more complex aromatic profile which differentiated them from the others. Biochemical features of breads mainly reflected those of the corresponding sourdough, whereas nutritional (protein digestibility and glycemic index) and technological (texture profile, colorimetric coordinates) features were hardly affected by the propagation method. Thus, investigation on the effect of the variation of the ecological determinants within the same propagation methods and their role in the definition of sourdough potential could be the subject of further studies.
Hide AbstractSupplementation with electrically conductive materials unlocks lactic acid conversion into volatile fatty acids during cheese whey fermentation.
Petitta, C., Tucci, M., Daghio, M., Capelli, C., Viti, C., Adessi, A., di Palma, L., Viggi, C. C. & Aulenta, F. (2025). Journal of Environmental Chemical Engineering, 13(4), 117197.
Cheese whey (CW), a by-product of the dairy industry, poses environmental challenges due to its high organic load and substantial production volumes. Dark fermentation (DF) offers a promising biological approach to valorizing CW by converting its carbohydrate-rich organic matter into valuable products such as organic acids, alcohols, and hydrogen. This study investigated the application of electrically conductive materials (ECMs)-specifically magnetite, biochar, and graphite-to enhance CW fermentation and increase the production of high-value volatile fatty acids (VFAs). Batch fermentation experiments revealed that incorporating ECMs significantly influenced the DF process. Notably, VFA production, particularly propionic acid, was markedly enhanced. In unamended control microcosms, CW fermentation led to an almost complete conversion of carbohydrates into lactic acid. Among the ECMs tested, magnetite had the greatest impact, increasing total VFA concentrations to 45.3 ± 5.9 g COD/L-a 22.5-fold improvement over the control. The addition of ECMs promoted the growth and enrichment of microorganisms capable of lactic acid reduction into propionic acid, such as Clostridiaceae and Propionibacteraceae, while also altering the microbial community and electron flow dynamics. This resulted in a significant increase in acetic acid production, which was over five times higher in ECM-amended treatments compared to controls. ECMs likely facilitated the disposal of excess reducing power, possibly via direct interspecies electron transfer (DIET), which further enhanced lactic acid conversion to propionic acid. From an environmental perspective, this study offers a sustainable solution for managing CW, reducing its environmental impact by converting it into valuable biochemicals. From an industrial standpoint, the enhanced production of VFAs, particularly propionic and acetic acids, presents a pathway to generate precursors for bio-based polymers, food additives, and other high-value applications.
Hide AbstractDevelopment, optimization and integrated characterization of rice-based yogurt alternatives enriched with roasted and non-roasted sprouted barley flour.
Caponio, M., Verni, M., Tlais, A. Z. A., Longo, E., Pontonio, E., Di Cagno, R. & Rizzello, C. G. (2025). Current Research in Food Science, 10, 101059.
Plant-based yogurt substitutes (“gurts”), whose market growth is steadily increasing, have emerged as a promising option to promote more sustainable diets and food systems, especially when produced with locally sourced or low-input crops like barley. In this study, a novel gurt made with rice (10%) and sprouted barley (5%), was designed. Four lactic acid bacteria strains, Levilactobacillus brevis AM7, Leuconostoc pseudomesenteroides DSM20193, Lactiplantibacillus plantarum 18S9 and H64, were used as starters for making prototypes. Although with some differences in their acidification kinetics and proteolysis, all the strains adapted to the matrix. Then the formulation and production process were optimized. The use of sprouted barley, compared to raw flours, provided a content of amino acids 9-fold higher, further increased (up to 35%) by the fermentation, and a more complex aroma profile characterized by the presence of furans and aldehydes. However, the high amylolytic activity in sprouted barley interfered with starch gelatinization decreasing the viscosity of the products from 3.3 to 0.08 Pa*s. To overcome this challenge and obtain a creamy and spoonable product, sprouted barley flour was roasted, deactivating the enzymes and conferring a nutty and toasted flavor to the gurts due to the presence of pyrazines. The stability of the key biochemical and microbiological parameters during refrigerated storage was also assessed. Hence, plant-based gurts made with sprouted barley, emerge as a sustainable and health-promoting substitute to traditional dairy yogurts.
Hide AbstractEnrichment of the nutritional value of pea flour milling fractions through fermentation.
Saula, T., Cigić, B., Jamnik, P., Cigić, I. K., Ulrih, N. P., Pozrl, T. & Marolt, G. (2025). Food chemistry, 476, 143303.
In this work, pea flour and two milling fractions obtained by industrial-scale air classification were characterized and fermented by Lactiplantibacillus plantarum to increase their nutritional value. Scanning electron microscopy and chemical analysis revealed major differences in the morphology and composition of the flours. Protein-rich (43.7%) fraction exhibits a few-fold higher mineral, spermidine (290 μg/g), but also a higher phytate (20.4 mg/g) content compared to starch-rich fraction. Flour type and inoculum majorly influenced the composition of the fermented product. In spontaneously fermented flours, biogenic amines accumulated up to 6.6 mg/g, which was the main drawback besides the large variations between batches, as confirmed by metagenomic analysis. Higher contents of lactic acid, free amino groups formed by proteolysis and gamma-aminobutyric acid were determined in inoculated fermentations of protein rich fraction, whereas a higher relative bioavailability of minerals was found in the inoculated starch-rich fraction, as the phytate content was reduced by 42%.
Hide AbstractMixed Culture of Yeast and Lactic Acid Bacteria for Low-Temperature Fermentation of Wheat Dough.
Liszkowska, W., Motyl, I., Pielech-Przybylska, K., Dziekońska-Kubczak, U. & Berłowska, J. (2025). Molecules, 30(1), 112.
There is growing interest in low-temperature food processing. In the baking industry, low-temperature fermentation improves the production of natural aroma compounds, which have a positive impact on the sensory profile of the final product. The aim of this study was to develop a yeast-lactic acid bacteria starter culture that effectively ferments wheat dough at a temperature of 15°C. The microorganisms were selected based on their enzymatic activity and ability to grow at low temperature. The fermentation activity of the yeast and mixed cultures was assessed enzymatically. The biosynthesis of volatile organic compounds was quantified using the HS-GC-MS technique. Samples fermented by S. cerevisiae D3 were characterized by the highest concentration of volatile organic compounds, especially esters. The addition of lactic acid bacteria increased not only the biosynthesis of volatile organic compounds but also the productivity of carbon dioxide during dough fermentation. Based on both dough expansion and the profile of volatile organic compounds, a mixed culture of S. cerevisiae D3 and L. brevis B46 was selected as the most effective starter for low-temperature fermentation.
Hide AbstractMolecular characterization of exopolysaccharide from Periweissella beninensis LMG 25373T and technological properties in plant-based food production.
Montemurro, M., Verni, M., Fanelli, F., Wang, Y., Maina, H. N., Torreggiani, A., Lamminaho, E., Coda, R., Fusco, V. & Rizzello, C. G. (2025). Food Research International, 201, 115537.
Periweissella beninensis LMG 25373T, belonging to the recently established Periweissella genus, exhibits unique motility and high adhesion capabilities, indicating significant probiotic potential, including resilience under simulated gastrointestinal conditions. This study demonstrates for the first time that P. beninensis LMG 25373^T produces a dextran-type exopolysaccharide (EPS) with a distinctive high degree of branching (approximately 71% of α-(1 → 6)-linkages and 29% α-(1 → 3)-linkages). Growth performance, acidification, and proteolytic activity were investigated in various plant-based substrates (lentil, chickpea, and rice flours water soluble extracts and semi-liquid mixtures), in comparison with the well-characterized lactic acid bacteria strains Leuconostoc pseudomesenteroides DSM 20193 (EPS-producing) and Lacticaseibacillus rhamnosus GG (probiotic). The strain displayed effective pro-technological properties, especially in gelatinized and non-gelatinized legume-based substrates, achieving EPS synthesis levels of up to 1.3 g/100 g and 2.7 ± 0.2 g/100 g, respectively. When used as a starter for a plant-based yogurt-type (“gurt”) prototype, compared to the control, P. beninensis LMG 25373T produced a substantial increase in viscosity which remained stable during refrigerated storage, confirming the role of its unique structure pattern as a hydrocolloid. Furthermore, the strain demonstrated high viability throughout storage, an essential trait for probiotic food applications.
Hide AbstractRatio of L-(+)-and D-(−)-Lactic Acids Produced by Enterococcus faecalis Changes Depending on the Culture pH.
Matsunaga, K. & Komatsu, Y. (2024). Microbiology Research, 15(4), 2703-2712.
Enterococcus faecalis (E. faecalis) has been associated with the specific production of L-(+)-lactic acid. However, in this study, D-(−)-lactic acid production by E. faecalis was observed under specific pH conditions. E. faecalis PR31 exhibited a significant amount of D-(−)-lactic acid under a stirring culture in MRS broth at pH 4.5, 5.8, and 6.0, and the contents of D-(−)-lactic acid were 45.1, 35.9, and 36.2%, respectively. When the cell suspension prepared at a pH of 6.0 was reacted with L-(+)- or D-(−)-lactic acid, D-(−)- or L-(+)-lactic acid was produced, respectively, in a time- and dose-dependent manner. Therefore, this phenomenon of D-(−)-lactic acid production in PR31 was suggested to be due to the activation of the larA gene encoding lactate racemase that is present in PR31. However, even in the E. faecalis-type strain NBRC 100480, which contains neither larA nor vanH, encoding D-(−)-lactate dehydrogenase VanH, D-(−)-lactic acid was also produced at specific pH values. Therefore, the production of D-(−)-lactic acid in NBRC 100480 was thought to occur not via the activation of larA. The biological significance of D-(−)-lactic acid production in E. faecalis depending on the pH and the detailed underlying mechanism, including whether it is the same in PR31 and NBRC 100480, remain to be elucidated in future studies.
Hide AbstractUnveiling 14 novel 2-hydroxy acid racemization and epimerization reactions in the lactate racemase superfamily.
Urdiain-Arraiza, J., Vandenberghe, A., Dimitrova, G. & Desguin, B. (2024). Journal of Biological Chemistry, 301 (1), 108069.
2-hydroxy acids are organic carboxylic acids ubiquitous in the living world and are important building blocks in organic synthesis. Recently, the lactate racemase (LarA) superfamily, a diverse superfamily of 2-hydroxy acid racemases and epimerases using the nickel-pincer nucleotide (NPN) cofactor, has been uncovered. In this study, we performed a taxonomic analysis of the LarA superfamily, showing the distribution of lactate racemase homologs (LarAHs) sequences across the three domains of life. Subsequently, we overexpressed and purified nine LarAHs and investigated their biochemical properties and substrate specificities. We show that LarAHs from the lactate racemases group are more promiscuous than previously thought, with some members showing high specificity towards glycerate or 2-hydroxybutyrate. In other phylogenetic groups, we identified a new malate racemase and 2-hydroxyglutarate racemase, as well as a new 2-gluconate epimerase from an eukaryotic organism. We show that some LarAHs are able to isomerize up to 16 different substrates, mostly 2-hydroxy acids with hydrophobic side chains, thereby identifying 14 novel 2-hydroxy acid racemization and epimerization reactions catalyzed by LarAHs. These include the racemization of glycerate, 2-hydroxybutyrate, 2,4-dihydroxybutyrate, 2-hydroxyvalerate, 2-hydroxycaproate, 2,3-dihydroxyisovalérate, 2-hydroxy-3,3-dimethylbutyrate, 3-(4-hydroxyphenyl)lactate, 2-hydroxy-4-phenylbutyrate, and 2-hydroxy-4-oxo-4-phenylbutyrate. Additionally, we observed the C2-epimerization of all 2,3-dihydroxybutyrate stereoisomers (4-deoxy-DL-threonate and 4-deoxy-DL-erythronate) and the C2-epimerization of D-arabinarate epimers. Finally, through comparative analysis of Alphafold structural predictions, we identified key residues likely involved in substrate specificity and predicted the function of half of the LarAHs from the LarA superfamily. In conclusion, this study widely expands the scope of substrates isomerized by NPN-dependent enzymes.
Hide AbstractDextran-enriched pea-based ingredient from a combined enzymatic and fermentative bioprocessing. Design of an innovative plant-based spread.
Perri, G., Difonzo, G., Wang, Y., Verni, M., Caponio, G. R., Coda, R., Blandino, M. & Pontonio, E. (2024). Future Foods, 10, 100502.
In this study a plant-based spread was developed using dextran-enriched ingredients derived from pea flours, supplemented with defatted durum wheat germ and almond flour. Optimization of fermentation with Leuconostoc pseudomesenteroides DSM 20193, both with and without enzymatic hydrolysis, aimed to enhance exopolysaccharide production and the nutritional value of pea flours. Best results were achieved through enzymatic hydrolysis with Veron PS protease followed by fermentation at 25°C, resulting in elevated dextran levels and increased peptides and total free amino acid concentration in green and yellow pea-based ingredients. The yellow pea-based ingredient was selected for the final plant-based spread formulation, blended at 35% w/w, with 45% w/w defatted durum wheat germ, and 20% w/w almond flour. The resultant spread exhibited elastic and solid-like characteristics like milk-based spreadable cheese and yogurt, boasting 'high protein' (12.49 g/100g) and 'high fiber' (11.01 g/100g) designations. It maintained chemical, biochemical, and microbiological stability over a 10-day shelf-life under refrigerated conditions. Sensory evaluation confirmed the acceptability of the plant-based spread (PBS), highlighting a well-balanced aroma and a grainy, adhesive texture. This research underscores the potential of an integrated approach utilizing food-grade enzymes and fermentation for the in-situ production of dextran to create innovative, clean label, and plant-based foods.
Hide AbstractElectrified liquid–liquid interface strategy for sensing lactic acid in buttermilk extract.
Sudalaimani, S., Esokkiya, A., Kumar, K. S. & Giribabu, K. (2025). Food Chemistry, 463, 141493.
Lactic acid (LA) serves as a freshness marker in certain foods. In the present work, electrified interfaces of different nature (i.e., liquid-liquid and liquid-organogel) have been developed for the quantification of LA. Electrochemical sensing of LA at the liquid-organogel interface revealed that adsorptive stripping voltammetry, with a preconcentration time of 500 s offered better sensitivity. Electroanalytical ability of LA under optimized conditions displayed a detection limit of 0.97 μM and 0.71 μM with sensitivity of 2.84 nA μM−1 and 3.59 nA μM−1 for liquid-liquid and liquid-organogel interfaces respectively. Quantification of LA using the developed methodology has been demonstrated in buttermilk as the real matrix. Analysis demonstrate that electrified liquid-liquid and liquid-organogel interfaces are promising approach for sensing LA in buttermilk extract.
Hide AbstractIsolation and characterization of Bifidobacterium spp. from breast milk with different human milk oligosaccharides utilization and anti-inflammatory capacity.
Ma, X., Mo, J., Shi, L., Cheng, Y., Feng, J., Qin, J., Su, W., Lv, J., Li, S., Li, Q. & Han, B. (2024). Food Research International, 196, 115092.
Breast milk is the best source of nutrition for infants. Human milk oligosaccharides (HMOs) and the corresponding HMOs-consuming Bifidobacterium positively influence infant health. This study aims to isolate and characterize Bifidobacterium from breast milk of healthy Chinese mothers, identifying the most efficacious strains for inclusion in simulated maternal milk formulas. Nine Bifidobacterium strains (two of B. breve and seven of B. infantis) were isolated, exhibiting a broad spectrum of probiotic potential. This included tolerance to simulated infant gastrointestinal conditions, notable adhesion, antibacterial, antioxidant activities, and HMOs utilization ability. Lacto-N-Tetraose (LNT) is preferred in early growth among Bifidobacterium isolates. B. breve showed a preference for LNT, whereas B. infantis showed a preference for fucosylated HMOs, and displayed reduced utilization of sialylated HMOs. They also exhibited robust safety profiles, including no hemolytic activity, an appropriate D/L lactate-producing ratio, and non-toxicity in an acute oral toxicity assay on mice. It is noteworthy that B. breve N-90, O-147, B. infantis O-161 and R-1 exhibited anti-inflammatory effects in LPS-induced RAW 264.7 cells. Specifically, a notable reduction in TNF-α levels was observed in pre-treatment, while a decrease in IL-1β and IL-6 levels in co-treatment. B. breve N-90 and B. infantis R-1 were identified finally as promising probiotic candidates. Their whole-genome sequencing analysis confirmed presence of functional genes associated with gastrointestinal colonization, antioxidation, and glycoside hydrolase activity on HMOs. The annotation for antibiotic resistance and virulence genes concurred with phenotypes, further validating the safety. Breast milk is a good source for Bifidobacteria isolation, while Bifidobacteria utilize HMOs in a strain-dependent manner. The two selected strains, B. breve N-90 and B. infantis R-1, are potential candidates for inclusion in simulated maternal milk formulas and deserved further in vivo investigation for their health-promoting effects.
Hide AbstractUnraveling the benefits of Bacillus subtilis DSM 29784 poultry probiotic through its secreted metabolites: an in vitro approach.
Vieco-Saiz, N., Prévéraud, D. P., Pinloche, E., Morat, A., Govindin, P., Blottière, H. M., Matthieu, E., Devillard, E. & Consuegra, J. (2024). Microbiology Spectrum, 12(11), e00177-24.
The probiotic Bacillus subtilis 29784 (Bs29784) sustains chicken's intestinal health, enhancing animal resilience and performance through the production of the bioactive metabolites hypoxanthine (HPX), niacin (NIA), and pantothenate (PTH). Here, using enterocyte in vitro models, we determine the functional link between these metabolites and the three pillars of intestinal resilience: immune response, intestinal barrier, and microbiota. We evaluated in vitro the capacity of Bs29784 vegetative cells, spores, and metabolites to modulate global immune regulators (using HT-29-NF-κB and HT-29-AP-1 reporter cells), intestinal integrity (HT-29-MUC2 reporter cells and Caco-2 cells), and cytokine production (Caco-2 cells). Finally, we simulated intestinal fermentations using chicken's intestinal contents as inocula to determine the effect of Bs29784 metabolites on the microbiota and their fermentation profile. Bs29784 vegetative cells reduced the inflammatory response more effectively than spores, indicating that their benefit is linked to metabolic activity. To assess this hypothesis, we studied Bs29784 metabolites individually. The results showed that each metabolite had different beneficial effects. PTH and NIA reduced the activation of the pro-inflammatory pathways AP-1 and NF-κB. HPX upregulated mucin production by enhancing MUC2 expression. HPX, NIA, and PTH increased cell proliferation. PTH and HPX increased epithelial resilience to an inflammatory challenge by limiting permeability increase. In cecal fermentations, NIA increased acetate, HPX increased butyrate, whereas PTH increased acetate, butyrate, and propionate. In ileal fermentations, PTH increased butyrate. All molecules modulated microbiota, explaining the different fermentation patterns. Altogether, we show that Bs29784 influences intestinal health by acting on the three lines of resilience via its secreted metabolites.
Hide AbstractEnhancing High-Pressure Bacterial Inactivation by Modified Atmosphere Packaging: Effect of Exposure Time and Cooked Ham Formulation.
Serra-Castelló, C., Jofré, A. & Bover-Cid, S. (2024). Food and Bioprocess Technology, 1-10.
High-pressure processing (HPP) is a non-thermal preservation technology that can be applied as a control measure to inactivate pathogens and spoilage microorganisms once RTE meat products are packaged in a convenient format. HPP efficacy highly depends on product characteristics, but the impact of the sodium-reduced formulations and the effect of packaging atmosphere are scarcely known. The aim of the present work was to assess the effect of standard and sodium-reduced formulations from two different brands (A, B) under different packaging (vacuum and modified atmosphere packaging (MAP)) on the HPP inactivation kinetics of Listeria monocytogenes and spoilage lactic acid bacteria in cooked ham. Slices of cooked ham with standard and sodium-reduced formulations were inoculated with L. monocytogenes CTC1034 and Latilactobacillus sakei CTC746 (slime producer), packaged in vacuum and MAP (CO2:N2, 20:80), and pressurized (400 MPa/0–15 min) after 1 h (vacuum, MAP) or 24 h (MAP-exposed). Parameters of HPP inactivation kinetics were estimated by fitting the Weibull model to log reduction data. Results showed that the efficacy of HPP in sodium-reduced cooked hams tended to decrease compared to standard formulations, being the difference statistically significant for L. sakei. For L. monocytogenes, a significant enhancing effect of MAP was observed when HPP was applied just after packaging (1 h, MAP) of cooked ham of brand A. In the case of L. sakei, the inactivation by HPP was only enhanced in MAP-exposed samples. Therefore, the use of HPP as a control measure must be applied through a product-oriented approach considering the type of packaging and the time period between packaging and HPP.
Hide Abstract