60 assays (manual) / 600 assays (microplate) / 600 assays (auto-analyser)
| Content: | 60 assays (manual) / 600 assays (microplate) / 600 assays (auto-analyser) |
| Shipping Temperature: | Ambient |
| Storage Temperature: |
Short term stability: 2-8oC, Long term stability: See individual component labels |
| Stability: | > 2 years under recommended storage conditions |
| Analyte: | Ethanol |
| Assay Format: | Spectrophotometer, Microplate, Auto-analyser |
| Detection Method: | Absorbance |
| Wavelength (nm): | 340 |
| Signal Response: | Increase |
| Linear Range: | 0.25 to 12 µg of ethanol per assay |
| Limit of Detection: | 0.093 mg/L |
| Reaction Time (min): | ~ 5 min |
| Application examples: | Wine, beer, cider, alcoholic fruit juices, spirits, liqueurs, low-alcoholic / non-alcoholic beverages, pickles, fruit and fruit juice, chocolate products, vinegar, jam, bread and bakery products, honey, soy sauce, dairy products, cosmetics, pharmaceuticals and other materials (e.g. biological cultures, samples, etc.). |
| Method recognition: | Methods based on this principle have been accepted by AOAC (AOAC Method 2019.08, First Action), IFU, EBC Method 9.3.1, MEBAK and ASBC Method Beer 4-F |
The Ethanol test kit is a simple, reliable and accurate method for the measurement and analysis of ethanol in beverages and foodstuffs.
Note for Content: The number of manual tests per kit can be doubled if all volumes are halved. This can be readily accommodated using the MegaQuantTM Wave Spectrophotometer (D-MQWAVE).
View our full range of alcohol assay kits.

- Extended cofactors stability. Dissolved cofactors stable for > 1 year at 4oC.
- Simple format – aldehyde dehydrogenase supplied as stable suspension
- Very competitive price (cost per test)
- All reagents stable for > 2 years after preparation
- Rapid reaction
- Mega-Calc™ software tool is available from our website for hassle-free raw data processing
- Standard included
- Suitable for manual, microplate and auto-analyser formats
Determination of ethanol concentration in Kombucha beverages: Single-laboratory validation of an enzymatic method, First Action Method 2019.08.
Ivory, R., Delaney, E., Mangan & McCleary, B. V. (2020). Journal of AOAC International, qsaa122.
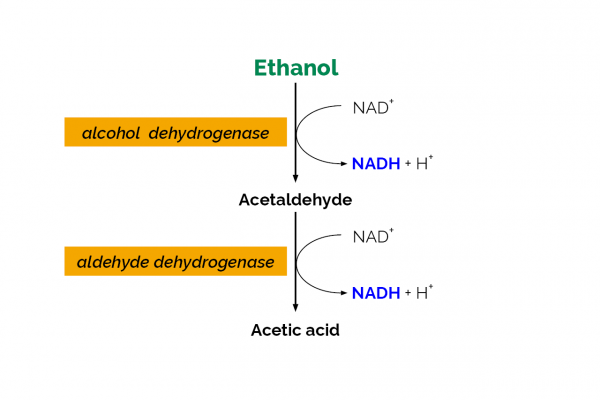
The Ethanol Assay Kit is an enzymatic test kit developed by Megazyme for the determination of ethanol in a variety of samples. The kit has been validated in a single laboratory for use with Kombucha fermented drinks, fruit juices and low-alcohol beer samples. The commercially available Ethanol Assay Kit (Megazyme catalogue no. K-ETOH) contains all components required for the analysis. Quantification is based on the oxidation of ethanol to acetaldehyde by alcohol dehydrogenase and further oxidation of acetaldehyde by acetaldehyde dehydrogenase with conversion of NAD+ to NADH. The single laboratory validation (SLV) outlined in this document was performed on a sample set of eight different commercial Kombucha products purchased in Ireland, a set of five Cerilliant aqueous ethanol solutions, two BCR low-alcohol beer reference materials, two alcohol-free beer samples and two fruit juice samples against SMPR 2016.001 (1). Parameters examined during the validation included Working range, Selectivity, Limit of Detection (LOD), Limit of Quantification (LOQ), Trueness (bias), Precision (reproducibility and repeatability), Robustness and Stability.
Hide AbstractMegazyme “advanced” wine test kits general characteristics and validation.
Charnock, S. J., McCleary, B. V., Daverede, C. & Gallant, P. (2006). Reveue des Oenologues, 120, 1-5.
Many of the enzymatic test kits are official methods of prestigious organisations such as the Association of Official Analytical Chemicals (AOAC) and the American Association of Cereal Chemists (AACC) in response to the interest from oenologists. Megazyme decided to use its long history of enzymatic bio-analysis to make a significant contribution to the wine industry, by the development of a range of advanced enzymatic test kits. This task has now been successfully completed through the strategic and comprehensive process of identifying limitations of existing enzymatic bio-analysis test kits where they occurred, and then using advanced techniques, such as molecular biology (photo 1), to rapidly overcome them. Novel test kits have also been developed for analytes of emerging interest to the oenologist, such as yeast available nitrogen (YAN; see pages 2-3 of issue 117 article), or where previously enzymes were simply either not available, or were too expensive to employ, such as for D-mannitol analysis.
Hide AbstractGrape and wine analysis: Oenologists to exploit advanced test kits.
Charnock, S. C. & McCleary, B. V. (2005). Revue des Enology, 117, 1-5.
It is without doubt that testing plays a pivotal role throughout the whole of the vinification process. To produce the best possible quality wine and to minimise process problems such as “stuck” fermentation or troublesome infections, it is now recognised that if possible testing should begin prior to harvesting of the grapes and continue through to bottling. Traditional methods of wine analysis are often expensive, time consuming, require either elaborate equipment or specialist expertise and frequently lack accuracy. However, enzymatic bio-analysis enables the accurate measurement of the vast majority of analytes of interest to the wine maker, using just one piece of apparatus, the spectrophotometer (see previous issue No. 116 for a detailed technical review). Grape juice and wine are amenable to enzymatic testing as being liquids they are homogenous, easy to manipulate, and can generally be analysed without any sample preparation.
Hide AbstractUnveiling the impact of traditional sourdough propagation methods on the microbiological, biochemical, sensory, and technological properties of sourdough and bread: a comprehensive first study.
Pontonio, E., Perri, G., Calasso, M., Celano, G., Verni, M. & Rizzello, C. G. (2025). Applied Food Research, 5(1), 101037.
In the last thirty years, the factors driving the establishment and composition of the sourdough biota have been deeply studied. Nevertheless, to date, no study has ever evaluated the biochemical and microbial dynamics of sourdoughs propagated using the different traditional methods integrated into procedural back-slopping practices worldwide. A mature type I sourdough was propagated for 10 days according to four managing conditions (Milanese, In Water, Free and Piedmontese) entailing incubations in a jute sack, submerged in water, in a jar or a combination of them. Sourdoughs obtained under the different conditions (and corresponding breads) were extensively characterized. When processing parameters modified the sourdough environment, the microbial community changed. In the first days of propagation Fructilactobacillus sanfranciscensis was the main dominant species regardless of the type of propagation, remaining present in all sourdoughs, especially those maintained in a jar. Differences among the propagation methods emerged from the biochemical analysis. Sourdoughs propagated in water exhibited higher titratable acidity, mainly due to the acetic acid produced, and were characterized by a more complex aromatic profile which differentiated them from the others. Biochemical features of breads mainly reflected those of the corresponding sourdough, whereas nutritional (protein digestibility and glycemic index) and technological (texture profile, colorimetric coordinates) features were hardly affected by the propagation method. Thus, investigation on the effect of the variation of the ecological determinants within the same propagation methods and their role in the definition of sourdough potential could be the subject of further studies.
Hide AbstractValorisation of insect infested sweet sorghum reeds towards production of a fermented beverage.
Makopa, T. P., Semumu, T., Gaaipone, M. T., Masemola, T., Ramchuran, S., Vrhovsek, U. & Zhou, N. (2025). BMC microbiology, 25(1), 1-19.
Sweet sorghum variety (Sorghum bicolour (L)) commonly known as sweet reeds, Ntšhe, in Setswana, is a valuable cash crop mostly for small scale farmers in Botswana and other southern African countries. These reeds are widely consumed as a delicacy and contribute significantly to food security, employment, and rural incomes. However, infestations by the larval stages of Chilo partellus (stem borer moths) lead to substantial economic losses, as consumers reject worm-infested reeds. To mitigate these losses, valorisation of condemned sweet reeds is attractive. Here, we took advantage of our understanding of yeast-insect interactions to isolate yeasts associated with larval stages of the stem borer moths and investigated their potential for use in the production of an alcoholic sweet sorghum beverage. We report the isolation of thirty-two yeast strains from the larvae and assessed their ability to ferment the simplest sugar, glucose, a constituent of the sweet sorghum juice. Out of the selected yeasts, a subset of fourteen strains belonging to Hanseniaspora and Candida genera were further characterised based on their capacity to ferment more sugars found in sweet sorghum juice. We further assessed the isolates for the ability to tolerate brewing/fermentation-associated stresses and production of complex aroma profiles towards the use of sweet sorghum juice as a sole feedstock to produce a commercial beverage. Our findings suggest that yeast-insect interactions offer a promising approach for converting rejected sweet sorghum stalks into a novel alcoholic beverage, adding economic value to an otherwise discarded resource.
Hide AbstractMixed Culture of Yeast and Lactic Acid Bacteria for Low-Temperature Fermentation of Wheat Dough.
Liszkowska, W., Motyl, I., Pielech-Przybylska, K., Dziekońska-Kubczak, U. & Berłowska, J. (2025). Molecules, 30(1), 112.
There is growing interest in low-temperature food processing. In the baking industry, low-temperature fermentation improves the production of natural aroma compounds, which have a positive impact on the sensory profile of the final product. The aim of this study was to develop a yeast-lactic acid bacteria starter culture that effectively ferments wheat dough at a temperature of 15°C. The microorganisms were selected based on their enzymatic activity and ability to grow at low temperature. The fermentation activity of the yeast and mixed cultures was assessed enzymatically. The biosynthesis of volatile organic compounds was quantified using the HS-GC-MS technique. Samples fermented by S. cerevisiae D3 were characterized by the highest concentration of volatile organic compounds, especially esters. The addition of lactic acid bacteria increased not only the biosynthesis of volatile organic compounds but also the productivity of carbon dioxide during dough fermentation. Based on both dough expansion and the profile of volatile organic compounds, a mixed culture of S. cerevisiae D3 and L. brevis B46 was selected as the most effective starter for low-temperature fermentation.
Hide AbstractTargeting protein aggregation using a cocoa-bean shell extract to reduce α-synuclein toxicity in models of Parkinson's disease.
Tripodi, F., Lambiase, A., Moukham, H., Spandri, G., Brioschi, M., Falletta, E., D'Urzo, A.,Vai, M., Abbiati, F., Pagliari, S., Salvo, A., Spano, M., Campone, L., Labra, M. & Coccetti, P. (2024). Current Research in Food Science, 9, 100888.
Neurodegenerative diseases are among the major challenges in modern medicine, due to the progressive aging of the world population. Among these, Parkinson's disease (PD) affects 10 million people worldwide and is associated with the aggregation of the presynaptic protein α-synuclein (α-syn). Here we use two different PD models, yeast cells and neuroblastoma cells overexpressing α-syn, to investigate the protective effect of an extract from the cocoa shell, which is a by-product of the roasting process of cocoa beans. The LC-ESI-qTOF-MS and NMR analyses allow the identification of amino acids (including the essential ones), organic acids, lactate and glycerol, confirming also the presence of the two methylxanthines, namely caffeine and theobromine. The present study demonstrates that the supplementation with the cocoa bean shell extract (CBSE) strongly improves the longevity of yeast cells expressing α-syn, reducing the level of reactive oxygen species, activating autophagy and reducing the intracellular protein aggresomes. These anti-aggregation properties are confirmed also in neuroblastoma cells, where CBSE treatment leads to activation of AMPK kinase and to a significant reduction of toxic α-syn oligomers. Results obtained by surface plasmon resonance (SPR) assay highlights that CBSE binds α-syn protein in a concentration-dependent manner, supporting its inhibitory role on the amyloid aggregation of α-syn. These findings suggest that the supplementation with CBSE in the form of nutraceuticals may represent a promising way to prevent neurodegenerative diseases associated with α-syn aggregation.
Hide AbstractEngineering new-to-nature biochemical conversions by combining fermentative metabolism with respiratory modules.
Schulz-Mirbach, H., Krüsemann, J. L., Andreadaki, T., Nerlich, J. N., Mavrothalassiti, E., Boecker, S., Schneider, P., Weresow, M., Abdelwahab, O., Paczia, N., Dronsella, B., Erb, T. J., Bar-Even, A., Klamt, S. & Lindner, S. N. (2024). Nature Communications, 15(1), 6725.
Anaerobic microbial fermentations provide high product yields and are a cornerstone of industrial bio-based processes. However, the need for redox balancing limits the array of fermentable substrate-product combinations. To overcome this limitation, here we design an aerobic fermentative metabolism that allows the introduction of selected respiratory modules. These can use oxygen to re-balance otherwise unbalanced fermentations, hence achieving controlled respiro-fermentative growth. Following this design, we engineer and characterize an obligate fermentative Escherichia coli strain that aerobically ferments glucose to stoichiometric amounts of lactate. We then re-integrate the quinone-dependent glycerol 3-phosphate dehydrogenase and demonstrate glycerol fermentation to lactate while selectively transferring the surplus of electrons to the respiratory chain. To showcase the potential of this fermentation mode, we direct fermentative flux from glycerol towards isobutanol production. In summary, our design permits using oxygen to selectively re-balance fermentations. This concept is an advance freeing highly efficient microbial fermentation from the limitations imposed by traditional redox balancing.
Hide AbstractLethal metabolism of Candida albicans respiratory mutants.
Kane, D. L., Burke Jr, B., Diaz, M., Wolf, C. & Fonzi, W. A. (2024). Plos one, 19(4), e0300630.
The destructive impact of fungi in agriculture and animal and human health, coincident with increases in antifungal resistance, underscores the need for new and alternative drug targets to counteract these trends. Cellular metabolism relies on many intermediates with intrinsic toxicity and promiscuous enzymatic activity generates others. Fuller knowledge of these toxic entities and their generation may offer opportunities of antifungal development. From this perspective our observation of media-conditional lethal metabolism in respiratory mutants of the opportunistic fungal pathogen Candida albicans was of interest. C. albicans mutants defective in NADH:ubiquinone oxidoreductase (Complex I of the electron transport chain) exhibit normal growth in synthetic complete medium. In YPD medium, however, the mutants grow normally until early stationary phase whereupon a dramatic loss of viability occurs. Upwards of 90% of cells die over the subsequent four to six hours with a loss of membrane integrity. The extent of cell death was proportional to the amount of BactoPeptone, and to a lesser extent, the amount of yeast extract. YPD medium conditioned by growth of the mutant was toxic to wild-type cells indicating mutant metabolism established a toxic milieu in the media. Conditioned media contained a volatile component that contributed to toxicity, but only in the presence of a component of BactoPeptone. Fractionation experiments revealed purine nucleosides or bases as the synergistic component. GC-mass spectrometry analysis revealed acetal (1,1-diethoxyethane) as the active volatile. This previously unreported and lethal synergistic interaction of acetal and purines suggests a hitherto unrecognized toxic metabolism potentially exploitable in the search for antifungal targets.
Hide AbstractUsing ethanol as postharvest treatment to increase polyphenols and anthocyanins in wine grape.
Margherita, M., Gianmarco, A., Anna, M., Roberto, F., Serena, F., Milena, P., Isabella, T., Fabio, M. & Andrea, B. (2024). Heliyon, 10(4).
Red wine grapes are qualitatively evaluated for their content in polyphenols and anthocyanins. Due to certain conditions (weather, latitude, temperature), the concentration of these compounds may be not at the right level for reaching a high-quality wine, thus postharvest technologies can be operated as a remediation strategy. Ethanol is a secondary volatile metabolite and its application has been demonstrated to delay fruit ripening, to reduce decay, and to increase secondary metabolites. The present study investigates the effects of ethanol post-harvest application on wine grapes' metabolism and composition. Red wine grapes (Vitis Vinifera L. cv Aglianico) were exposed to different ethanol doses (0.25, 0.5, or 1 mL L-1) for 12, 24, or 36 h. Ethanol increased sugar concentration, malic acid, free amino nitrogen, polyphenols, and anthocyanins. Particularly, anthocyanins reached an average value of 1820 mg/L in treated samples versus the 1200 mg/L of control grapes already after 12 h whatever the concentration was. Moreover, the highest concentration of ethanol modified berry metabolism shifting from aerobic to anaerobic one. Obtained results suggest that 12 h of ethanol postharvest treatment could be an interesting solution to improve anthocyanins in wine grapes, especially when the quality is not as good as expected.
Hide AbstractEvaluating the Effect of Adding Selected Herbs, Spices, and Fruits to Fermented Olympus Mountain Tea (Sideritis scardica) Kombucha Sweetened with Thyme Honey: Assessment of Physicochemical and Functional Properties.
Geraris Kartelias, I., Panagiotakopoulos, I., Nasopoulou, C. & Karantonis, H. C. (2024). Beverages, 10(1), 9.
This study examined the effects of adding herbs, spices, and fruits into fermented Olympus Mountain tea (Sideritis scardica) kombucha using thyme honey as a sweetener. This study evaluated how these additions affected the tea’s physical, chemical, and functional characteristics. Two different enrichments were proposed: a “Golden Mountain tea and honey Kombucha” (KG) with fresh ginger, turmeric powder, and lemon zest and juice and a “Red Mountain tea and honey Kombucha” (KR) with dried hibiscus calyces, rose petals, and lavender blossoms. In KR, the levels of vitamin C increased from 33.2 ± 2.7 to 48.4 ± 4.5. Additionally, the levels of calcium increased from 31.0 ± 1.2 to 55.7 ± 1.2, while the levels of potassium practically doubled from 64.7 ± 0.6 to 115.7 ± 2.5. An increased potassium concentration was observed in KG, and ionic iron was found for the first time after both enrichments. The total phenolic and flavonoid contents, along with antioxidant capacity, as assessed by the ABTS and DPPH methods, were found to be substantially enhanced in KR. In KG, the total phenolic content increased, together with antioxidant activity, as assessed by ABTS. Enrichment with hibiscus calyces, rose petals, and lavender blossoms significantly increased inhibitory effects against α-amylase, α-glucosidase, acetylcholinesterase, and butyrylcholinesterase. On the other hand, enrichment with ginger, turmeric, and lemon zest and juice decreased inhibitory effects against α-glucosidase and increased those against α-amylase, acetylcholinesterase, and butyrylcholinesterase. KR had the strongest enzyme-inhibiting activity, with its α-glucosidase-inhibiting activity increased by approximately 18 times. Therefore, enrichment with selected herbs, spices, and fruits can transform fermented Olympus Mountain tea kombucha sweetened with honey into a novel beverage with enhanced functional properties.
Hide AbstractGrowth-coupled anaerobic production of isobutanol from glucose in minimal medium with Escherichia coli.
Boecker, S., Schulze, P. & Klamt, S. (2023). Biotechnology for Biofuels and Bioproducts, 16(1), 148.
Background: The microbial production of isobutanol holds promise to become a sustainable alternative to fossil-based synthesis routes for this important chemical. Escherichia coli has been considered as one production host, however, due to redox imbalance, growth-coupled anaerobic production of isobutanol from glucose in E. coli is only possible if complex media additives or small amounts of oxygen are provided. These strategies have a negative impact on product yield, productivity, reproducibility, and production costs. Results: In this study, we propose a strategy based on acetate as co-substrate for resolving the redox imbalance. We constructed the E. coli background strain SB001 (ΔldhA ΔfrdA ΔpflB) with blocked pathways from glucose to alternative fermentation products but with an enabled pathway for acetate uptake and subsequent conversion to ethanol via acetyl-CoA. This strain, if equipped with the isobutanol production plasmid pIBA4, showed robust exponential growth (µ = 0.05 h−1) under anaerobic conditions in minimal glucose medium supplemented with small amounts of acetate. In small-scale batch cultivations, the strain reached a glucose uptake rate of 4.8 mmol gDW−1 h−1, a titer of 74 mM and 89% of the theoretical maximal isobutanol/glucose yield, while secreting only small amounts of ethanol synthesized from acetate. Furthermore, we show that the strain keeps a high metabolic activity also in a pulsed fed-batch bioreactor cultivation, even if cell growth is impaired by the accumulation of isobutanol in the medium. Conclusions: This study showcases the beneficial utilization of acetate as a co-substrate and redox sink to facilitate growth-coupled production of isobutanol under anaerobic conditions. This approach holds potential for other applications with different production hosts and/or substrate–product combinations.
Hide AbstractThe marula and elephant intoxication myth: assessing the biodiversity of fermenting yeasts associated with marula fruits (Sclerocarya birrea).
Makopa, T. P., Modikwe, G., Vrhovsek, U., Lotti, C., Sampaio, J. P. & Zhou, N. (2023). FEMS microbes, 4, xtad018.
The inebriation of wild African elephants from eating the ripened and rotting fruit of the marula tree is a persistent myth in Southern Africa. However, the yeasts responsible for alcoholic fermentation to intoxicate the elephants remain poorly documented. In this study, we considered Botswana, a country with the world's largest population of wild elephants, and where the marula tree is indigenous, abundant and protected, to assess the occurrence and biodiversity of yeasts with a potential to ferment and subsequently inebriate the wild elephants. We collected marula fruits from over a stretch of 800 km in Botswana and isolated 106 yeast strains representing 24 yeast species. Over 93% of these isolates, typically known to ferment simple sugars and produce ethanol comprising of high ethanol producers belonging to Saccharomyces, Brettanomyces, and Pichia, and intermediate ethanol producers Wickerhamomyces, Zygotorulaspora, Candida, Hanseniaspora, and Kluyveromyces. Fermentation of marula juice revealed convincing fermentative and aromatic bouquet credentials to suggest the potential to influence foraging behaviour and inebriate elephants in nature. There is insufficient evidence to refute the aforementioned myth. This work serves as the first work towards understanding the biodiversity marula associated yeasts to debunk the myth or approve the facts.
Hide AbstractThe phenotype and genotype of fermentative prokaryotes.
Hackmann, T. J. & Zhang, B. (2023). Science Advances, 9(39), eadg8687.
Fermentation is a type of metabolism pervasive in oxygen-deprived environments. Despite its importance, we know little about the range and traits of organisms that carry out this metabolism. Our study addresses this gap with a comprehensive analysis of the phenotype and genotype of fermentative prokaryotes. We assembled a dataset with phenotypic records of 8350 organisms plus 4355 genomes and 13.6 million genes. Our analysis reveals fermentation is both widespread (in ~30% of prokaryotes) and complex (forming ~300 combinations of metabolites). Furthermore, it points to previously uncharacterized proteins involved in this metabolism. Previous studies suggest that metabolic pathways for fermentation are well understood, but metabolic models built in our study show gaps in our knowledge. This study demonstrates the complexity of fermentation while showing that there is still much to learn about this metabolism. All resources in our study can be explored by the scientific community with an online, interactive tool.
Hide AbstractTreatment of food processing wastes for the production of medium chain fatty acids via chain elongation.
Battista, F., Zeni, A., Andreolli, M., Salvetti, E., Rizzioli, F., Lampis, S. & Bolzonella, D. (2024). Environmental Technology & Innovation, 33, 103453.
The production of medium chain fatty acids (MCFAs) through reverse β-oxidation was investigated both on synthetic and real substrates. From preliminary batch tests emerged that caproic acid was maximized under an acetate/ethanol molar ratio of 5:1 at neutral pH. This ratio was then adopted in different semi-continuous tests operating with different amounts of the two reactants. It emerged that the MCFAs yield reached the maximum level of 6.7% when the total molar substrate amount was around 40–45 mmol/d, while the process was inhibited for values higher than 400 mmol/d. Semi-continuous tests using real waste as substrates, namely food waste condensate, cheese whey, and winery wastewater, confirmed the results obtained with the synthetic substrates. Better performances were obtained when an adequate molar ratio of the acetate and the electron-donor compound was naturally present. Therefore, a MCFAs yield of 25% and 10.5% was obtained for condensate of food waste and acidic cheese whey, respectively. Regarding MCFAs composition, caproic acid was the dominant form but small concentrations of octanoic acid were also found in the tests where ethanol was the electron donor (synthetic substrates and food waste condensate). Octanoic acid was not produced in test where lactic acid represented the electron donor molecules (cheese whey). Condensate and synthetic samples were dominated by Pseudoclavibacter caeni with an abundance of 38.19% and 33.38% respectively, while Thomasclavelia (24.13%) and Caproiciproducens (11.68%) was the most representative genus in acidic cheese whey sample.
Hide Abstract