8 g
Prices exclude VAT
Available for shipping
| Content: | 8 g |
| Shipping Temperature: | Ambient |
| Storage Temperature: | Ambient |
| Physical Form: | Powder |
| Stability: | > 2 years under recommended storage conditions |
| CAS Number: | 11078-27-6 |
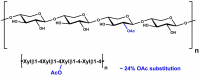
| Source: | Sugar-beet pulp |
| Purity: | > 95% |
| Monosaccharides (%): | Arabinose: Galactose: Rhamnose: Galacturonic acid: Other sugars = 69: 18.7: 1.4: 10.2: 0.7 |
| Main Chain Glycosidic Linkage: | α-1,5 |
| Substrate For (Enzyme): | endo-Arabinanase |
High purity Arabinan (Sugar Beet) for use in research, biochemical enzyme assays and in vitro diagnostic analysis.
See more of our high purity polysaccharides.
McCleary, B. V., McKie, V. A., Draga, A., Rooney, E., Mangan, D. & Larkin, J. (2015). Carbohydrate Research, 407, 79-96.
A range of α-L-arabinofuranosyl-(1-4)-β-D-xylo-oligosaccharides (AXOS) were produced by hydrolysis of wheat flour arabinoxylan (WAX) and acid debranched arabinoxylan (ADWAX), in the presence and absence of an AXH-d3 α-L-arabinofuranosidase, by several GH10 and GH11 β-xylanases. The structures of the oligosaccharides were characterised by GC-MS and NMR and by hydrolysis by a range of α-L-arabinofuranosidases and β-xylosidase. The AXOS were purified and used to characterise the action patterns of the specific α-L-arabinofuranosidases. These enzymes, in combination with either Cellvibrio mixtus or Neocallimastix patriciarum β -xylanase, were used to produce elevated levels of specific AXOS on hydrolysis of WAX, such as 32-α-L-Araf-(1-4)-β-D-xylobiose (A3X), 23-α-L-Araf-(1-4)-β-D-xylotriose (A2XX), 33-α-L-Araf-(1-4)-β-D-xylotriose (A3XX), 22-α-L-Araf-(1-4)-β-D-xylotriose (XA2X), 32-α-L-Araf (1-4)-β-D-xylotriose (XA3X), 23-α-L-Araf-(1-4)-β-D-xylotetraose (XA2XX), 33-α-L-Araf-(1-4)-β-D-xylotetraose (XA3XX), 23 ,33-di-α-L-Araf-(1-4)-β-D-xylotriose (A2+3XX), 23,33-di-α-L-Araf-(1-4)-β-D-xylotetraose (XA2+3XX), 24,34-di-α-L-Araf-(1-4)-β-D-xylopentaose (XA2+3XXX) and 33,34-di-α-L-Araf-(1-4)-β-D-xylopentaose (XA3A3XX), many of which have not previously been produced in sufficient quantities to allow their use as substrates in further enzymic studies. For A2,3XX, yields of approximately 16% of the starting material (wheat arabinoxylan) have been achieved. Mixtures of the α-L-arabinofuranosidases, with specific action on AXOS, have been combined with β-xylosidase and β-xylanase to obtain an optimal mixture for hydrolysis of arabinoxylan to L-arabinose and D-xylose.
Hide AbstractVerhertbruggen, Y., Marcus, S. E., Haeger, A., Verhoef, R., Schols, H. A., McCleary, B. V., McKee, L., Gilbert, H. J. & Knox, J. P. (2009). The Plant Journal, 59(3), 413-425.
Plant cell walls are constructed from a diversity of polysaccharide components. Molecular probes directed to structural elements of these polymers are required to assay polysaccharide structures in situ, and to determine polymer roles in the context of cell wall biology. Here, we report on the isolation and the characterization of three rat monoclonal antibodies that are directed to 1,5-linked arabinans and related polymers. LM13, LM16 and LM17, together with LM6, constitute a set of antibodies that can detect differing aspects of arabinan structures within cell walls. Each of these antibodies binds strongly to isolated sugar beet arabinan samples in ELISAs. Competitive-inhibition ELISAs indicate the antibodies bind differentially to arabinans with the binding of LM6 and LM17 being effectively inhibited by short oligoarabinosides. LM13 binds preferentially to longer oligoarabinosides, and its binding is highly sensitive to arabinanase action, indicating the recognition of a longer linearized arabinan epitope. In contrast, the binding of LM16 to branched arabinan and to cell walls is increased by arabinofuranosidase action. The presence of all epitopes can be differentially modulated in vitro using glycoside hydrolase family 43 and family 51 arabinofuranosidases. In addition, the LM16 epitope is sensitive to the action of β-galactosidase. Immunofluorescence microscopy indicates that the antibodies can be used to detect epitopes in cell walls, and that the four antibodies reveal complex patterns of epitope occurrence that vary between organs and species, and relate both to the probable processing of arabinan structural elements and the differing mechanical properties of cell walls.
Hide AbstractA key genetic factor governing arabinan utilization in the gut microbiome alleviates constipation.
Zhang, C., Yu, L., Ma, C., Jiang, S., Zhang, Y., Wang, S., Tian, F., Xue, Y., Zhao, J., Zhang, H., Liu, L., Chen, W., Huang, S., Zhang, J. & Zhai, Q. (2023). Cell Host & Microbe, 31(12), 1989-2006.
Impaired gastrointestinal motility is associated with gut dysbiosis. Probiotics, such as Bifidobacteria, can improve this bowel disorder; however, efficacy is strain-dependent. We determine that a genetic factor, the abfA cluster governing arabinan utilization, in Bifidobacterium longum impacts treatment efficacy against functional constipation (FC). In mice with FC, B. longum, but not an abfA mutant, improved gastrointestinal transit time, an affect that was dependent upon dietary arabinan. abfA genes were identified in other commensal bacteria, whose effects in ameliorating murine FC were similarly abfA-dependent. In a double-blind, randomized, placebo-controlled clinical trial, supplementation with abfA-cluster-carrying B. longum, but not an abfA-deficient strain, enriched arabinan-utilization residents, increased beneficial metabolites, and improved FC symptoms. Across human cohorts, abfA-cluster abundance can predict FC, and transplantation of abfA cluster-enriched human microbiota to FC-induced germ-free mice improved gut motility. Collectively, these findings demonstrate a role for microbial abfA cluster in ameliorating FC, establishing principles for genomics-directed probiotic therapies.
Hide AbstractArabinoxylan-based substrate preferences and predicted metabolic properties of Bifidobacterium longum subspecies as a basis to design differential media.
Calvete-Torre, I., Sabater, C., Delgado, S., Ruas-Madiedo, P., Rupérez-García, A., Montilla, A., Moreno. F. J., Margolles, A. & Ruiz, L. (2023). Food Research International, 167, 112711.
Arabinoxylan (AX) and arabinoxylo-oligosaccharides (AXOS) derived therefrom are emergent prebiotics with promising health promoting properties, likely linked to its capacity to foster beneficial species in the human gut. Bifidobacteria appear to be one taxa that is frequently promoted following AX or AXOS consumption, and that is known to establish metabolic cross-feeding networks with other beneficial commensal species. Therefore, probiotic bifidobacteria with the capability to metabolize AX-derived prebiotics represent interesting candidates to develop novel probiotic and synbiotic combinations with AX-based prebiotics. In this work we have deepen into the metabolic capabilities of bifidobacteria related to AX and AXOS metabolization through a combination of in silico an in vitro tools. Both approaches revealed that Bifidobacterium longum and, particularly, B. longum subsp. longum, appears as the better equipped to metabolize complex AX substrates, although other related subspecies such as B. longum subsp. infantis, also hold some machinery related to AXOS metabolization. This correlates to the growth profiles exhibited by representative strains of both subspecies in AX or AXOS enriched media. Based on these results, we formulated a differential carbohydrate free medium (CFM) supplemented with a combination of AX and AXOS that enabled to recover a wide diversity of Bifidobacterium species from complex fecal samples, while allowing easy discrimination of AX metabolising strains by the appearance of a precipitation halo. This new media represent an appealing alternative to isolate novel probiotic bifidobacteria, rapidly discriminating their capacity to metabolize structurally complex AX-derived prebiotics. This can be convenient to assist formulation of novel functional foods and supplements, including bifidobacterial species with capacity to metabolize AX-derived prebiotic ingredients.
Hide AbstractZobellia alginoliquefaciens sp. nov., a new flavobacteria isolated from the epibiota of the brown alga Ericaria zosteroides (C. Agardh) Molinari & Guiry 2020.
Barbeyron, T., Le Duff, N., Duchaud, E. & Thomas, F. (2023). BioRxiv, 2023-03.
Strain LLG6346-3.1T, isolated from the thallus of the brown alga Ericaria zosteroides collected in Mediterranean Sea near Bastia in Corsica, France, was characterized using a polyphasic method. Cells were Gram-stain-negative, strictly aerobic, non-flagellated, motile by gliding, rod-shaped and grew optimally at 30-33°C, at pH 8-8.5 and with 4-5 % NaCl. Strain LLG6346-3.1T used the seaweed polysaccharide alginic acid as sole carbon source which was vigorously liquefied. Phylogenetic analyses showed that the bacterium is affiliated to the genus Zobellia (family Flavobacteriaceae, class Flavobacteriia). Strain LLG6346-3.1T exhibited 16S rRNA gene sequence similarity values of 98.5 and 98.3 % to the type strains of Zobellia russellii and Zobellia roscoffensis respectively, and of 97.4-98.2 % to other species of the genus Zobellia. The DNA G+C content of strain LLG6346-3.1T was determined to be 38.28 mol%. Digital DNA-DNA hybridization predictions by the ANI and GGDC methods between strain LLG6346-3.1T and other members of the genus Zobellia showed values of 76-88 %, and below 37 %, respectively. The phenotypic, phylogenetic and genomic analyses show that strain LLG6346-3.1T is distinct from species of the genus Zobellia with validly published names and that it represents a novel species of the genus Zobellia, for which the name Zobellia alginoliquefaciens sp. nov. is proposed. The type strain is LLG6346-3.1T (RCC 7657T = LLG 32918T).
Hide AbstractGlycoside hydrolase family 2 exo-β-1, 6-galactosidase LpGal2 from Lactobacillus plantarum: Cloning, expression, and enzymatic characterization.
Zhang, X., Yu, G., Leng, J., Zhang, H., Zhou, Y., Yuan, Y. & Gao, J. (2021). Process Biochemistry, 102, 269-274.
Lactobacillus plantarum is a useful microorganism that metabolizes galactose-containing polysaccharides. Genome analysis has shown that L. plantarum contains four β-galactosidase-related genes. Here, we cloned the β-galactosidase gene that encodes the glycoside hydrolase family 2 (GH2) enzyme LpGal2. Recombinant LpGal2 (rLpGal2, 72 kDa) is a homodimer with maximal enzymatic activity at pH 7.0 and 50°C. Under these conditions, rLpGal2 hydrolyzes p-nitrophenyl-β-D-galactopyranoside (pNPβGal) with a specific activity of 2.16 × 10−3 U/mg and substrate specificity for β-1,6-galactobiose to produce D-Galactose. In addition, rLpGal2 can also hydrolyze β-1,6-galactan to D-Galactose, whereas other galactose-containing oligosaccharides and polysaccharides tested could not be hydrolyzed. This finding demonstrates that LpGal2 functions as an exo-β-1,6-galactosidase with narrow substrate specificity. To our knowledge, this is the first report of a β-galactosidase derived from L. plantarum with exo-β-1,6-galactosidase activity that has potential application for structure analysis of polysaccharides.
Hide AbstractLin, D., Lopez-Sanchez, P., Selway, N. & Gidley, M. J. (2018). Food Hydrocolloids, 79, 13-19.
The interactions between cellulose and pectin polysaccharides in primary plant cell walls are not fully understood, although several recent studies indicate that they might play an important role in wall properties. Studying polysaccharide interactions in planta is challenging, due to the complexity and heterogeneity of plant materials. Therefore, to investigate these interactions and the implications for the rheological properties of cell walls, we have taken a bottom-up approach in which cellulose/pectin composites are created either by adsorption of pectin polysaccharides (arabinan, galactan, homogalacturonan DE 69, homogalacturonan DE 33 and pectin DE 33) on cellulose-coated sensors in a quartz crystal microbalance with dissipation monitoring (QCM-D) or by incorporation of pectin during in vivo cellulose synthesis by Komagataeibacter bacteria. The viscoelastic behavior of the adsorbed layers was analyzed by applying the Voigt model to the QCM-D data, whilst the bulk viscoelastic properties of bacterial cellulose/pectin composites were studied by small amplitude oscillatory shear rheology. Our results show that all of the pectin polysaccharides studied have the ability to adsorb on the cellulose surfaces. The viscoelastic properties of the adsorbed layer varied depending on the substitution and degree of esterification of the pectin polysaccharides. Additionally, oscillatory rheology results showed that all bacterial cellulose-pectin composites had a gel nature (G′ > G″) with moduli varying in line with QCM-D determined viscoelasticity. Our interpretation of the results provides a better understanding of pectin-cellulose interactions and has implications for primary plant cell wall material properties.
Hide AbstractLarsbrink, J., Tuveng, T. R., Pope, P. B., Bulone, V., Eijsink, V. G., Brumer, H. & McKee, L. S. (2017). Journal of Proteomics, 156, 63-74.
Together with fungi, saprophytic bacteria are central to the decomposition and recycling of biomass in forest environments. The Bacteroidetes phylum is abundant in diverse habitats, and several species have been shown to be able to deconstruct a wide variety of complex carbohydrates. The genus Chitinophaga is often enriched in hotspots of plant and microbial biomass degradation. We present a proteomic assessment of the ability of Chitinophaga pinensis to grow on and degrade mannan polysaccharides, using an agarose plate-based method of protein collection to minimise contamination with exopolysaccharides and proteins from lysed cells, and to reflect the realistic setting of growth on a solid surface. We show that select Polysaccharide Utilisation Loci (PULs) are expressed in different growth conditions, and identify enzymes that may be involved in mannan degradation. By comparing proteomic and enzymatic profiles, we show evidence for the induced expression of enzymes and PULs in cells grown on mannan polysaccharides compared with cells grown on glucose. In addition, we show that the secretion of putative biomass-degrading enzymes during growth on glucose comprises a system for nutrient scavenging, which employs constitutively produced enzymes. Significance of this study: Chitinophaga pinensis belongs to a bacterial genus which is prominent in microbial communities in agricultural and forest environments, where plant and fungal biomass is intensively degraded. Such degradation is hugely significant in the recycling of carbon in the natural environment, and the enzymes responsible are of biotechnological relevance in emerging technologies involving the deconstruction of plant cell wall material. The bacterium has a comparatively large genome, which includes many uncharacterised carbohydrate-active enzymes. We present the first proteomic assessment of the biomass-degrading machinery of this species, focusing on mannan, an abundant plant cell wall hemicellulose. Our findings include the identification of several novel enzymes, which are promising targets for future biochemical characterisation. In addition, the data indicate the expression of specific Polysaccharide Utilisation Loci, induced in the presence of different growth substrates. We also highlight how a constitutive secretion of enzymes which deconstruct microbial biomass likely forms part of a nutrient scavenging process.
Hide AbstractThieme, N., Wu, V. W., Dietschmann, A., Salamov, A. A., Wang, M., Johnson, J., Singan, V. R., Grigoriev, I. V., Glass, N. L., Somerville, C. R., & Benz, J. P. (2017). Biotechnology for Biofuels, 10(1), 149.
Background: Pectin is an abundant component in many fruit and vegetable wastes and could therefore be an excellent resource for biorefinery, but is currently underutilized. Fungal pectinases already play a crucial role for industrial purposes, such as for foodstuff processing. However, the regulation of pectinase gene expression is still poorly understood. For an optimal utilization of plant biomass for biorefinery and biofuel production, a detailed analysis of the underlying regulatory mechanisms is warranted. In this study, we applied the genetic resources of the filamentous ascomycete species Neurospora crassa to screen for transcription factors that play a major role in pectinase induction. Results: The pectin degradation regulator-1 (PDR-1) was identified through a transcription factor mutant screen in N. crassa. The Δpdr-1 mutant exhibited a severe growth defect on pectin and all tested pectin-related poly- and monosaccharides. Biochemical as well as transcriptional analyses of WT and the Δpdr-1 mutant revealed that while PDR-1-mediated gene induction was dependent on the presence of L-rhamnose, it also strongly affected the degradation of the homogalacturonan backbone. The expression of the endo-polygalacturonase gh28-1 was greatly reduced in the Δpdr-1 mutant, while the expression levels of all pectate lyase genes increased. Moreover, a pdr-1 overexpression strain displayed substantially increased pectinase production. Promoter analysis of the PDR-1 regulon allowed refinement of the putative PDR-1 DNA-binding motif. Conclusions: PDR-1 is highly conserved in filamentous ascomycete fungi and is present in many pathogenic and industrially important fungi. Our data demonstrate that the function of PDR-1 in N. crassa combines features of two recently described transcription factors in Aspergillus niger (RhaR) and Botrytis cinerea (GaaR). The results presented in this study contribute to a broader understanding of how pectin degradation is orchestrated in filamentous fungi and how it could be manipulated for optimized pectinase production.
Hide AbstractTuncil, Y. E., Xiao, Y., Porter, N. T., Reuhs, B. L., Martens, E. C. & Hamaker, B. R. (2017). mBio, 8(5), e01068-17.
When presented with nutrient mixtures, several human gut Bacteroides species exhibit hierarchical utilization of glycans through a phenomenon that resembles catabolite repression. However, it is unclear how closely these observed physiological changes, often measured by altered transcription of glycan utilization genes, mirror actual glycan depletion. To understand the glycan prioritization strategies of two closely related human gut symbionts, Bacteroides ovatus and Bacteroides thetaiotaomicron, we performed a series of time course assays in which both species were individually grown in a medium with six different glycans that both species can degrade. Disappearance of the substrates and transcription of the corresponding polysaccharide utilization loci (PULs) were measured. Each species utilized some glycans before others, but with different priorities per species, providing insight into species-specific hierarchical preferences. In general, the presence of highly prioritized glycans repressed transcription of genes involved in utilizing lower-priority nutrients. However, transcriptional sensitivity to some glycans varied relative to the residual concentration in the medium, with some PULs that target high-priority substrates remaining highly expressed even after their target glycan had been mostly depleted. Coculturing of these organisms in the same mixture showed that the hierarchical orders generally remained the same, promoting stable coexistence. Polymer length was found to be a contributing factor for glycan utilization, thereby affecting its place in the hierarchy. Our findings not only elucidate how B. ovatus and B. thetaiotaomicron strategically access glycans to maintain coexistence but also support the prioritization of carbohydrate utilization based on carbohydrate structure, advancing our understanding of the relationships between diet and the gut microbiome.
Hide Abstract